Understanding Signal Transduction Networks
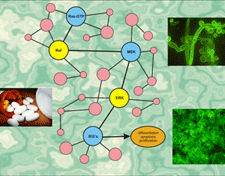
A visual representation of signal transduction networks.
All cells in the human body have the same genetic code, so how can a skin cell be so different from a liver cell? How are these differences regulated, and how can diseases, such as cancer, arise from misregulation? These questions and many more involve studying the time-dependent aspects (dynamics) of large complex networks composed of many elementary units interacting with each other in complex nonlinear fashion.
We are developing a general methodology for model-based identification and analysis of complex nonlinear signal transduction networks from incomplete and noisy observations. We are employing rigorous theoretical and computational techniques for estimating the structural and dynamic properties of such networks by state-of-the-art identification and model selection methodologies, and for studying network robustness via probabilistic sensitivity analysis. We use fundamental laws of physics (such as thermodynamics) to appropriately constrain the systems under consideration and focus our identification and analysis methodologies only on physically realizable networks. Sensitivity analysis is used from start to finish: in model reduction to ease inference, in experimental design to enhance scientific discovery, and in final system analysis for controlled pharmacological intervention.
Click Here for more about Dr. Goutsias' work >>
More about Computational Biology >>


