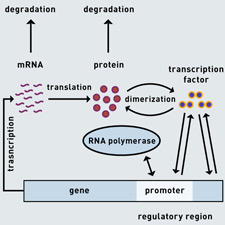
Modeling Gene Networks
To understand most cellular processes, one must understand how genetic information is processed. A formidable challenge is the dissection of gene regulatory networks to delineate how eukaryotic cells coordinate and govern patterns of gene expression that ultimately lead to a phenotype. Researchers at CIS are developing several approaches for modeling eukaryotic gene regulatory networks and for reverse engineering such networks from experimental observations.
Faculty: Geman, Goutsias, Younes
Featured Research: Understanding Signal Transduction Networks
Molecular Cancer Diagnosis
The mRNA counts in gene mircroarrays provide a global view of cellular activity by simultaneously recording the expression levels of thousands of genes. Nonetheless, there are technical and practical limitations in using these data for analyzing disease. Accurate statistical inference is difficult due to the small number of observations, typically tens, relative to the large number of genes, typically thousands. Moreover, conventional methods from machine learning lead to decisions which are usually very difficult to interpret in simple or biologically meaningful terms. Researchers at CIS and ICM are investigating a new approach to molecular classification based entirely on pairwise mRNA (or protein) comparisons ("relative expression reversals"). This provides simple classification rules which involve very few genes and generate specific hypotheses for follow-up studies.
Faculty: Geman